- Home /
- Help
Help
Search...
-
How to use the "TargetNet"?
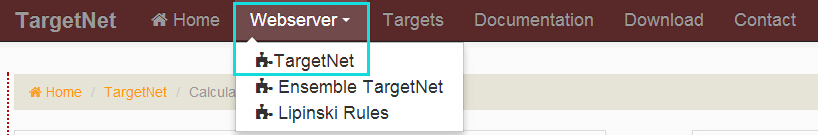
The TargetNet can only be accessed by selecting 'Webserver' link shown as the following figure.
Step 1: Users are required to submit a molecular structure or a molecular file with SMILES format.
Use SMILES to net the targets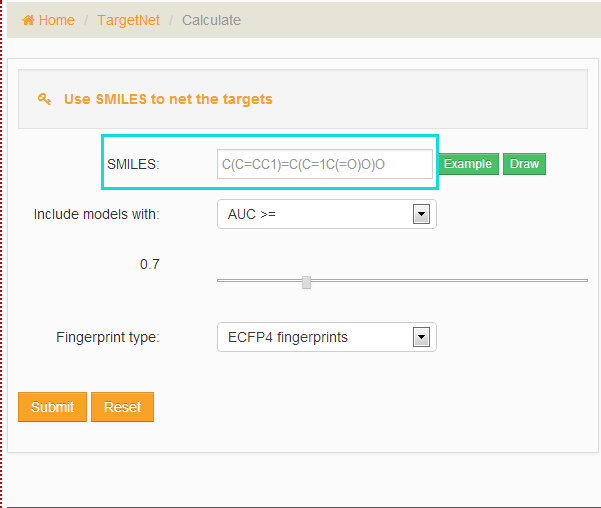
After entering three or more characters in the 'SMILES' field, choose the models and fingerprint type and then click the ‘Submit’ button. Users can click the ‘example’ button to load an example or use the JSDraw to Draw a molecule.
Upload file with molecular structures to net the targets
Choose a molecular file, models, fingerprint type and then click the ‘Submit’ button. Users can also click the ‘example’ button to download a molecular file for an example.
Step 2: Processing and results
The processing will last for a while and then users will be redirected to the result page. The result table contains "Details", "Uniprot_ID", "Protein", "Probability". "1" marks the "download" button, and users can choose different types of file to save the results. "2" marks the ‘per page’ button, and users can choose the number of items shown in the table. "3" marks the "search" button, and users can conveniently type in a keyword to look for a certain item in the results. "4" marks the ‘rank’ button, and users can click the button to get an increasing or decreasing order of the values. "5" marks the "details" button, and users can click here to get the detailed information of the target.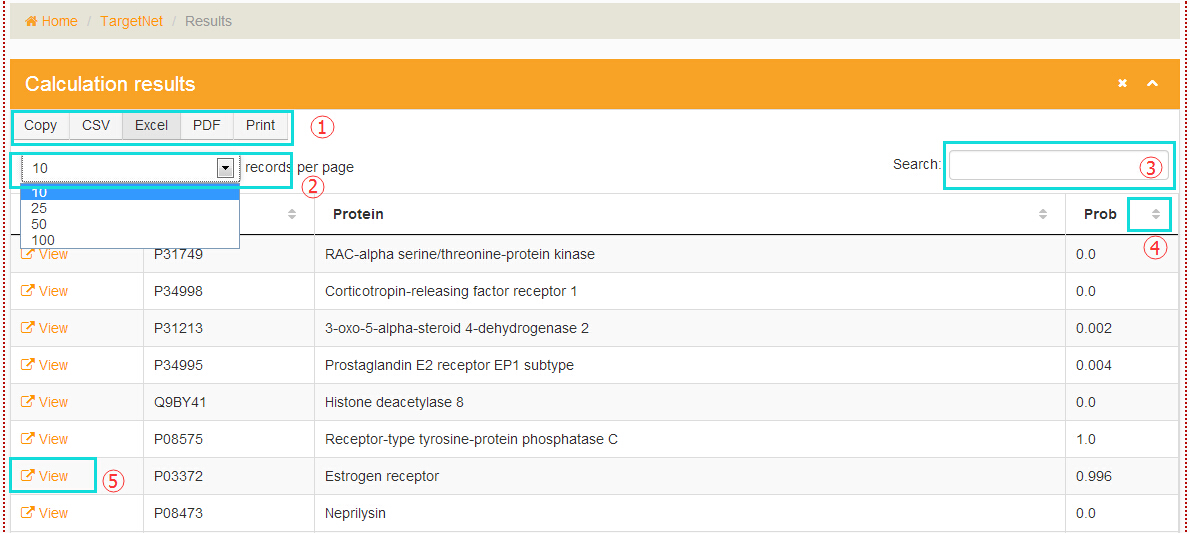
-
How to use the "Ensemble TargetNet" ?
Step 1: Users are required to submit a molecular structure or a molecular file with SMILES format.
Step 2: Processing and results.
These two steps are like the steps in "How to use the TargetNet". The difference is selecting the "Fingerprint types". Users can choose several kinds of fingerprints to access the ensemble results. -
How to calculate the "Lipinski Rule of Five"?
Step 1: Submitting a molecular structure or a molecular file with SMILES format.
Step 2: Processing and results.
The first step is like the step 1 in "How to use the TargetNet". In the step 2, the result table contains "Molecule", "TPSA", "MR", "Molecular weight", "HBD", "HBA1", "logP", "Lipinski Rule of Five" of the items. The last one is represented by the percentage of the entries that meet the criteria.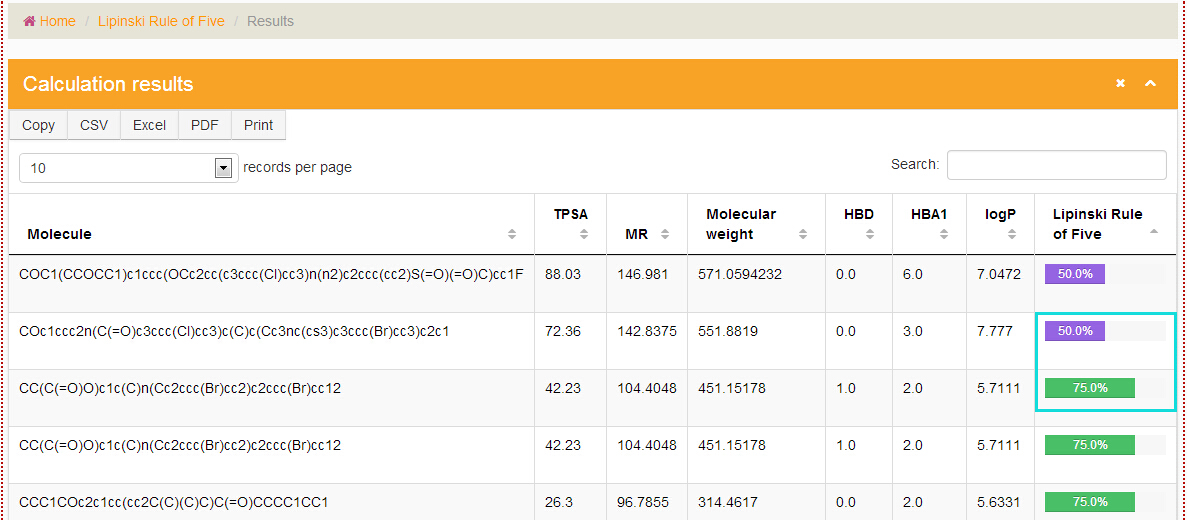
-
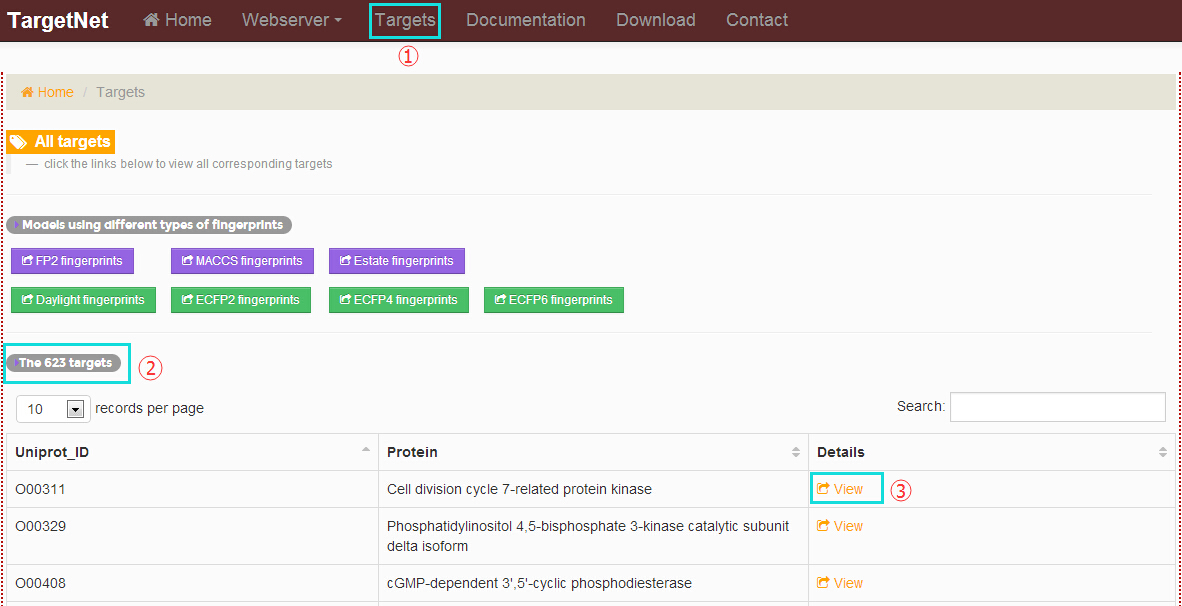
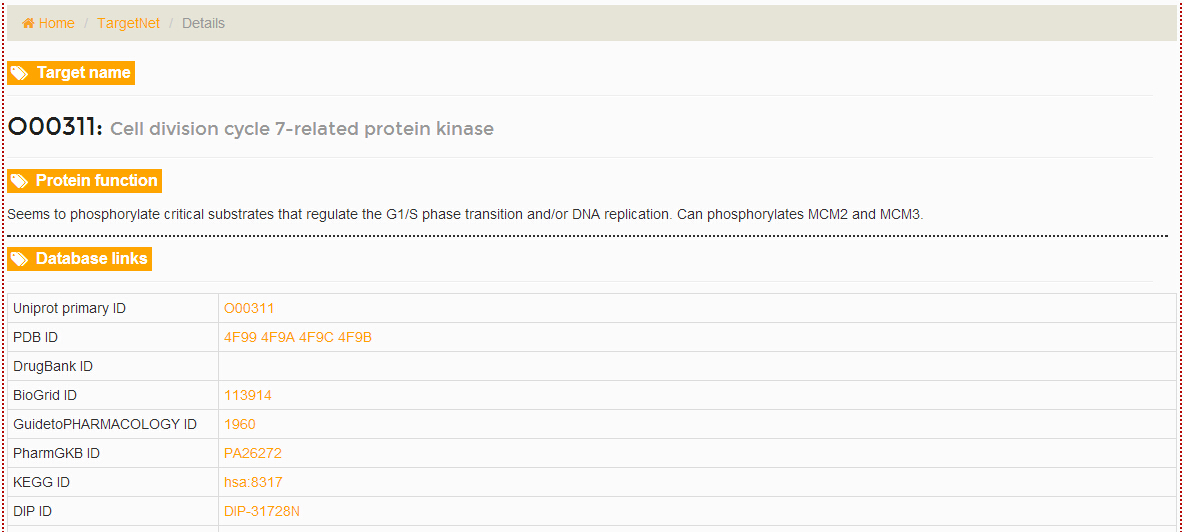
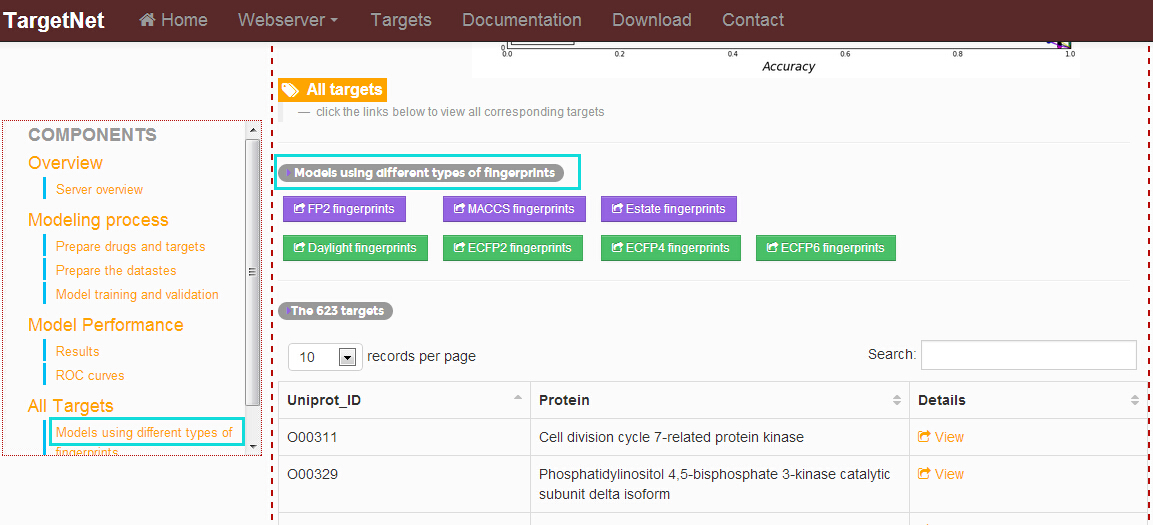
How to access the "All targets" and the details of each target?
Flowing the marks in the picture below users will get the ‘All targets’ and the details for each target.
-
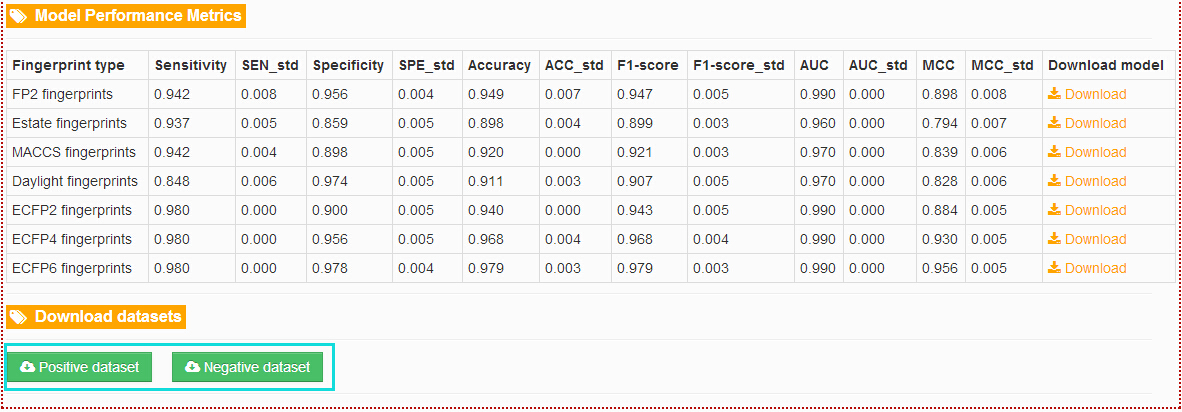
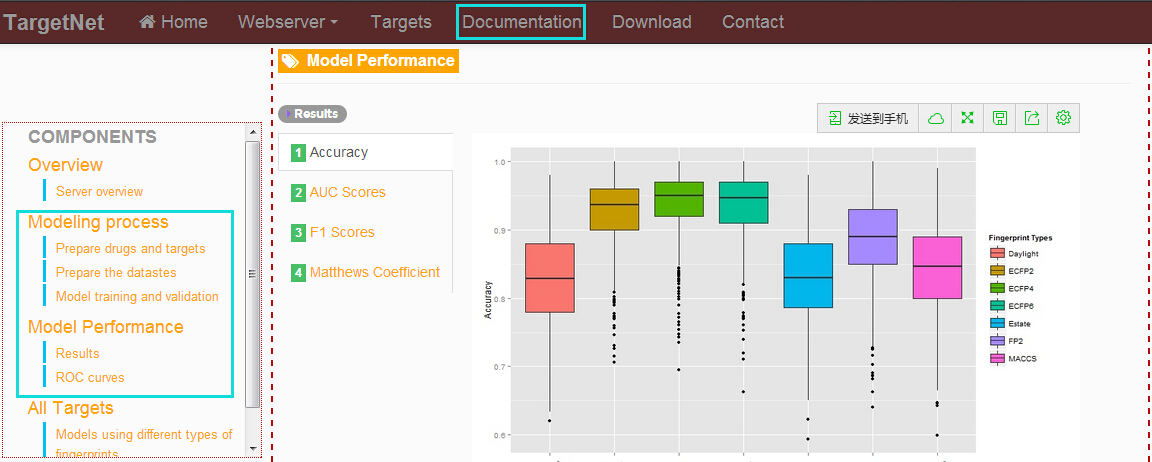
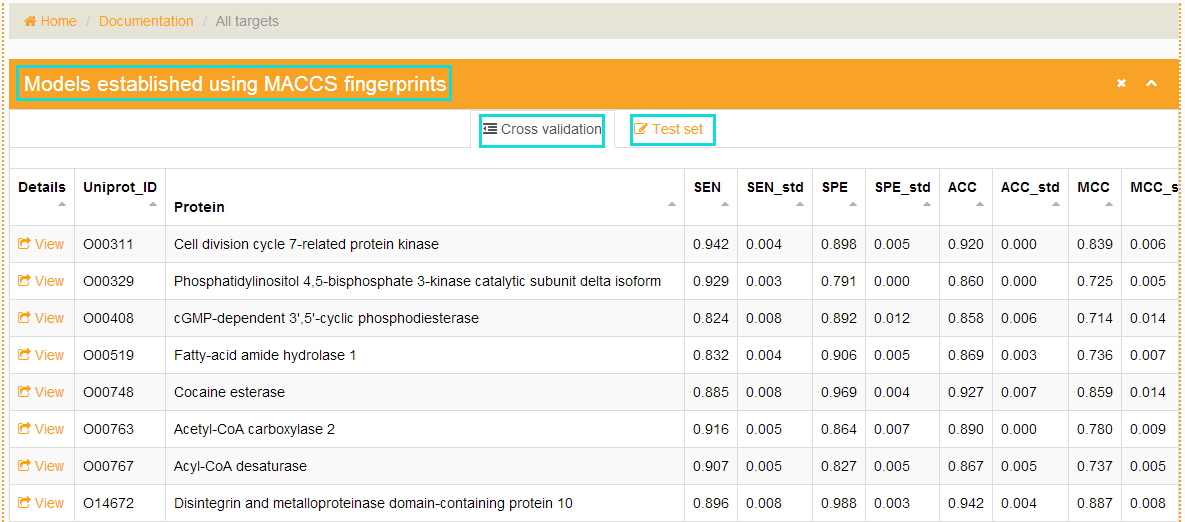
How to access the modeling process and models performance using different types of fingerprints?
In the "Documentation" section, click the links at the left navigation and users will get the modeling process and models performance using different types of fingerprints.
-
How to download the related resources used in TargetNet?
In the detail page of each target, users will get the "Database links", "Model Performance Metrics" and "Download datasets". Here, the positive dataset and negative dataset for the target can be downloaded. In the "Download" section at the top navigation provide all the related resources and all of them can be downloaded.